Active Learning for the Development of Molecular Neural Network Potentials
By Bart de Mooij (Dual Master in Computational Science and Chemistry UvA)
In a Nutshell
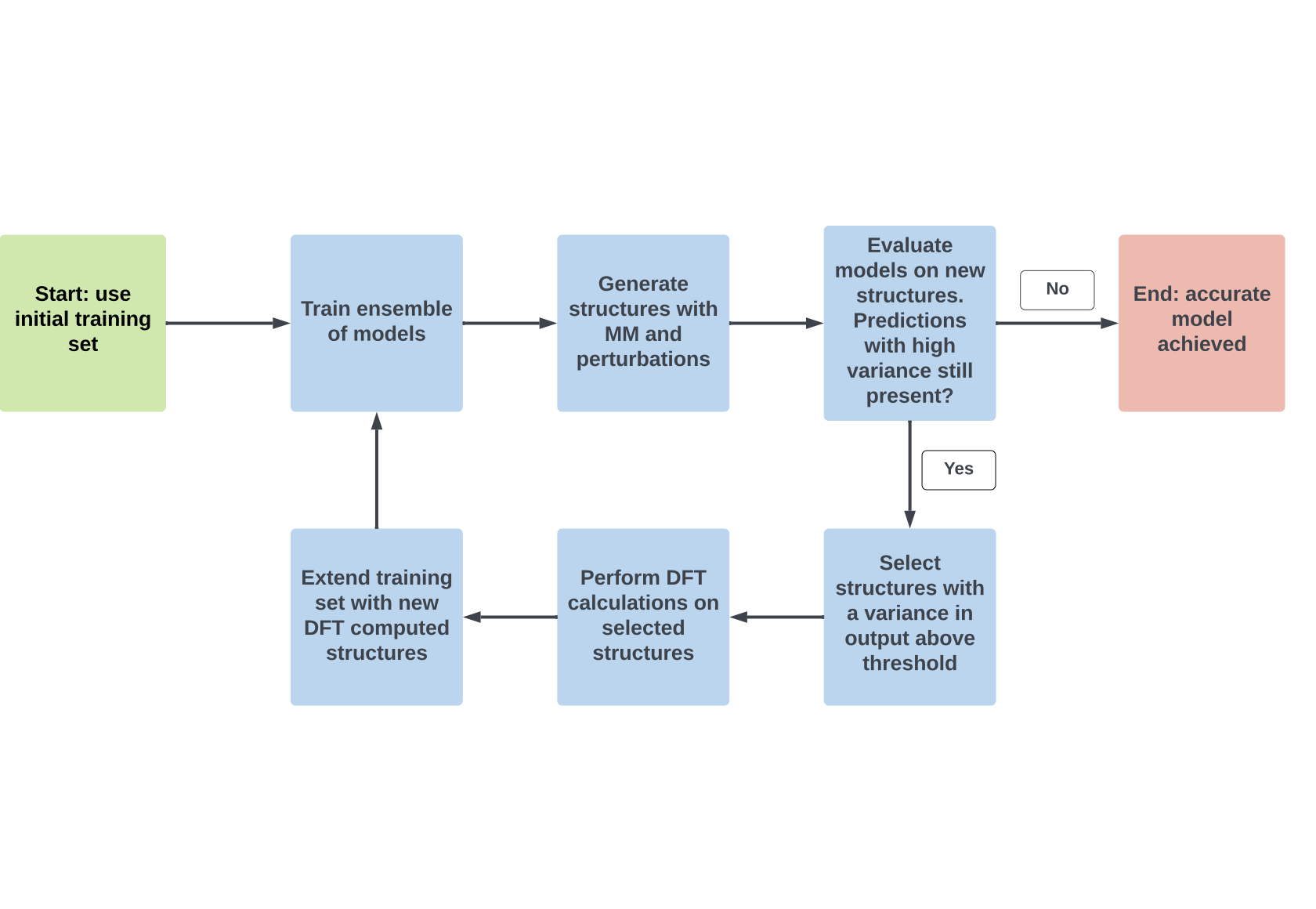
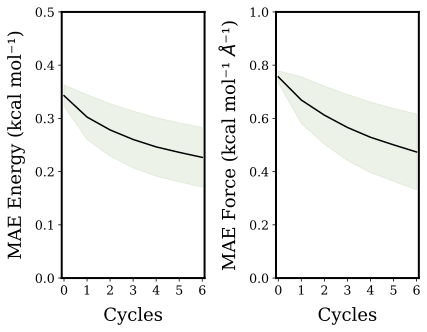
Machine Learning applications on molecular systems is a rapidly devel- oping field over recent years as it can retain quantum chemical accuracy while reducing computational cost. A recently proposed and relatively data efficient method is NequIP, an open-source package for creating, training, and using E(3)-equivariant machine learning interatomic potentials. The application of this method was tested against multiple molecular systems including an electrochemical system of formate dehydrogenation on fcc(110) copper. Formate dehydrogenation on copper is a chemically interesting sys- tem as it (1) includes covalent and metallic bonds, (2) charge transfer occurs between surface and adsorbate, (3) contains distinct molecular states affect- ing adsorbate’s bond lengths differently and (4) involves bond breaking. In addition, the formation of formate from carbon dioxide is important for sus- tainability purposes as the amount of carbon dioxide can be reduced while making a useful molecule. This research illustrated that ML on this cop- per formate system is more complicated than on smaller molecules. The developed ML models were able to accurately predict energies and forces, resulting in the prediction of MD trajectories with a comparable radial dis- tribution function to the underlying ground truth. This was equally so for geometry optimizations. Furthermore, considerable speed-ups were achieved for single point calculations using ML compared to DFT. Two fully auto- mated AL pipelines were developed applicable to either large readily avail- able data sets as well as when no data set was present. In the first scenario, a considerably smaller training set is necessary for accurate model generation as redundant structures in the data are typically omitted. For the second scenario, AL could generate its own reference data on the fly. Consequently, the computation of expensive quantum chemical calculation could be re- duced and limited to structures necessary to obtain an accurate ML model. Hence, both AL methods improve the efficiency of generating ML models.